our RESEARCH
RESEARCH THEMES
Our lab focuses on multimodal regulation, exploring how different regulatory mechanisms—genetic, cellular, metabolic, signalling, and environmental factors—integrate to control complex biological processes. We investigate how these layers of regulation work together to guide cell differentiation, tissue development, and metabolic responses, with a particular emphasis on their role in early mammalian embryonic development and focusing on mouse and human embryos. By studying how cellular signalling pathways, metabolic cues, and (post)transcriptional modifications coordinate, we aim to uncover the intricate processes driving cell fate decisions and morphogenesis.
To find out more about our lab’s focus and projects:
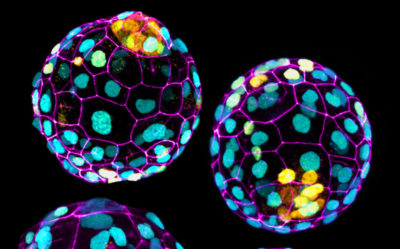
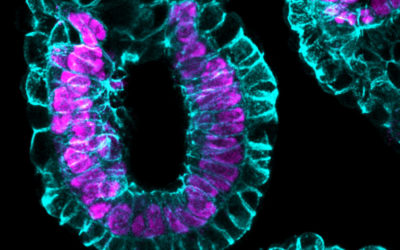
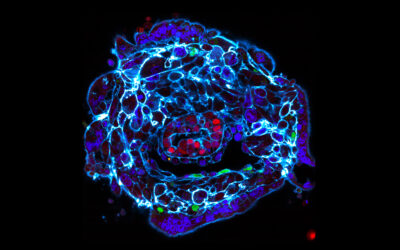
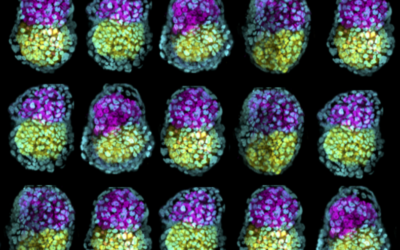
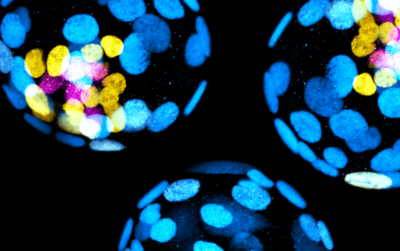
Modelling mammalian development in 3D
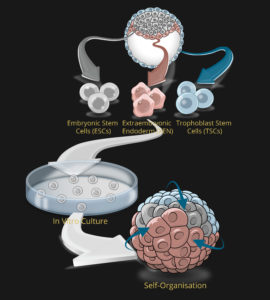
In order to investigate dynamic morphogenetic patterning at early embryogenesis, we exploit the self-organization properties of stem cells to model corresponding native lineages/tissues. We developed pioneering techniques to mimic early embryogenesis using mouse embryonic and extra-embryonic stem cells in vitro.
WHY IS THIS WORK IMPORTANT?
Stem cell–based model systems are important to study early embryogenesis for two main reasons:
(1) our models include not only embryonic but also extra-embryonic tissues, and therefore, recapitulates the architecture and gene expression signatures of the natural embryo, and
(2) formation does not require external signals; self-organization occurs organically through cellular interactions.
This system allows us to:
- capture and replicate early cell potency, programming, and pattern formation in vitro.
- use a bottom-up approach to piece together, step-by-step, the spatiotemporal events of embryogenesis.
- recreate the complex cellular organization and micro-environmental signals that define native tissues.
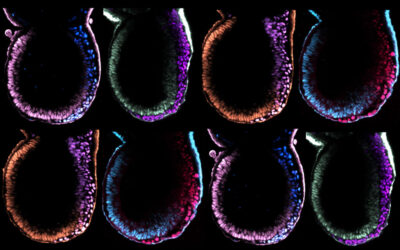
STUDYING DEVELOPMENTAL PROGRAMMING
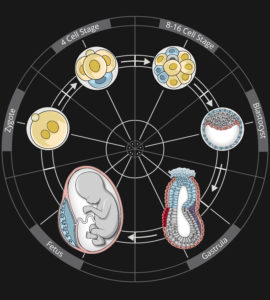
Upon implantation, mammalian embryos undergo profound morphological remodelling and establish vast cellular diversity initiated by the emergence of embryonic and placental progenitors. Concomitantly, pluripotency becomes established in the embryonic epiblast lineage, a developmental transition that precedes the formation of the primary germ layers, progenitors of all major organs and the embryo proper. Pluripotency is a transient, metastable cell state and exquisitely sensitive to extrinsic signals from the environment. Hence, environmental cues transmitted through pluripotency pathways have the potential to modify the default developmental trajectory defined by the genome and can result in permanent changes in the physiology, metabolism, transcriptome and epigenome in offspring, events that culminate in developmental programming.
We study precisely when and how regulative cellular and molecular mechanisms, guided by maternal nutritional imbalance, compromises embryo health, leading to adverse pregnancy outcomes.
WHY IS THIS WORK IMPORTANT?
Despite its central importance, presently there is a very scant mechanistic understanding of environmentally-mediated developmental reprogramming in early embryogenesis.
Our research helps to understand how events in early life shape later life morbidity risk; and provides translational insights into the developmental origins of disease. Our research translates into an understanding of the healthy development of embryos in the context of reproductive medicine and assisted reproductive technologies (ART).

LINKING METABOLISM TO PATTERNING
Studying environmental cues brings us specific interest in the interplay between metabolism and transcriptional regulation during embryonic patterning.
Recent studies showed that the cellular metabolic state is causative of the establishment and exit from pluripotency to initiate differentiation, not correlative. Our major objective is to understand how metabolism is mechanistically linked to spatiotemporal germ layer patterning through its bioenergetic and metabolic signalling functions.
A direct consequence of differential metabolic activity is the dynamic change in metabolite levels over time and space. In turn, cellular levels of intermediate metabolites modulate activities of chromatin-modifying enzymes, and therefore, metabolite levels contribute to the temporal control of gene expression which subsequently alters cell fate.
Through single-cell multi-omics, we map molecular trajectories of developmental adaptive plasticity during human embryonic patterning and aim to define transcriptomic changes that play an etiologic role in the downstream phenotypic, metabolic, and physiological consequences that lead to adverse pregnancy outcomes.
WHY IS THIS WORK IMPORTANT?
Establishing cellular diversity during the second week of development, when the embryo-to-gastrula transition occurs, is undoubtedly one of the most fundamental processes in an embryo’s life, and a major effector of pregnancy outcomes.
Our studies provide insights into the interconnections between cellular, metabolic, and molecular mechanisms that influence the phenotype of all successive cell types as they develop and differentiate.
